- Manipulating epigenetic mechanisms in neurologic genetic diseases
Angelman syndromes is a rare neurogenetic disease that is the textbook example of imprinting disorder. We are using artificial transcription factors to activate the epigenetically silenced gene in in the brain. This project is funded by the Foundation for Angelman Syndrome Therapeutics.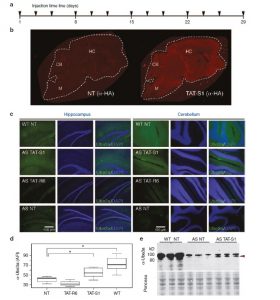 Reactivation of Ube3a in a mouse model of AS by TAT-S1 but not TAT-R6.
Reactivation of Ube3a in a mouse model of AS by TAT-S1 but not TAT-R6. - Functional genomics of non-coding elements
In collaboration with Peggy Farnham and the ENCODE consortium, we are using targetable nucleases and transcription factors based on zinc fingers, Transcription Activator-like Effector (TALE), and Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein (CRISPR/Cas) to disrupt non-coding genetic elements in the human genome to better understand their function. Our most recent efforts focus on creating epigenomic editing tools that can precisely manipulate epigenetic information at specific loci. Such tools can be used for the long-term control of gene expression for both research and therapeutic applications. - Genetic variation in health and disease
Several genetic variations (SNPs) have been associated with an increased risk of common complex disorders, such as colorectal cancer. In collaboration with Luis Carvajal-Carmona, we are using targetable nucleases identify causative SNPs and determine their mechanism of function. Our most recent efforts focus on creating tools that can precisely alter a single base pair at specific loci. Our approach overcomes the historic barrier of trying to study the affects of specific human mutations in a background of millions of other genetic differences between two individuals. - High-throughput investigations of CRISPR-DNA interactions
We continue to develop new methodologies for genome editing, such as methods to study off-target activity of CRISPRs and factors for targeted epigenetic modification. We employ methods of directed evolution for protein engineering and ChIP-seq and RNA-seq to examine the effects of tools on a genome-wide scale.
Zinc finger, TALE, CRISPR/Cas genome engineering and targeted gene regulation for applications in research and therapeutics, especially neurologic disorders.